Angle closure glaucoma (ACG) is not widely known to be a familial condition, yet the recent explosion of genetic data and large scale genome wide investigations have confirmed at least 13 genetic loci associated with ACG [1], and provided some insight into the clinical and pathobiological mechanisms of the disease.
Primary angle closure glaucoma (PACG) is defined as iridotrabecular contact with evidence of elevated intraocular pressure (IOP), or peripheral anterior synechiae (PAS), and glaucomatous optic neuropathy (GON), with a reproducible visual field defect [2]. PACG is often diagnosed at the extremes of phenotypes when there is advanced chronic visual loss, or acute angle closure (AAC) [3]. Understanding the genetic aetiology for PACG can help identify at-risk individuals at earlier stages of the disease.
Family history reporting
There are more people worldwide bilaterally blind from PACG than progressive open angle glaucoma (POAG) [4] but reported family history of PACG is less well studied. A clinic report from Brazil found 25% of their cases to have a reported family history of PACG [5]. A population survey in Harbin, Northeast China, found a positive family history to be a significant risk factor of PACG, with an odds ratio of 65 under univariate logistic regression, or odds ratio of 1.65 in the multivariate model [6]. Amerasinghe et al. found that the relative risk of narrow angles to first degree family members is seven times higher than the general population of Singaporean Chinese [7]. Kong et al. compared 332 PAC patients, 228 POAG and 193 controls in Shanghai, China to investigate a glaucoma family history, finding that a parent with PACG contributed most to the family history of glaucoma in the PAC, compared to POAG where siblings and children contributed more to the family history of glaucoma [8]. In India, Kavitha et al. found that there was a 13.6 times greater odds of angle-closure in siblings of PAC/G subjects compared to open angle subjects in a hospital setting [9]. A greater age effect was observed for glaucoma in PACG.
Accurate phenotyping is crucial
When Lowe et al. first described a series of families with angle-closure in Australia [10], little was known about the natural history of the disease. He noted that the rate of AAC increased as anterior chamber depth (ACDs) got shallower, but the co-inheritance of PACG in multiple family members was much less than angle-closure observed without glaucoma. He proposed a major environmental component to the disease mechanism, while emphasising that familial PACG was rare but definitely observed [11]. Subsequent groups, such as Spaeth et al., Tomlinson et al. and Sihota et al. all reported small ocular biometry to be prominent in familial disease [12-14]. However, all these reports had low rates of AAC.
Using the Spaeth gonioscopic grading system, iris convexity was found to be a common feature amongst affected family members [13], and more recently anterior segment imaging with ultrasound biomicroscopy (UBM) and anterior segment optical coherence tomography (AS-OCT) have allowed detailed, objective quantification of angle closure traits such as thicker irides, anteriorly rotated ciliary bodies, and iris vault to be studied. Figure 1 shows a high resolution AS-OCT scan of an affected individual with iridotrabecular contact. Three pedigrees with multiple affected individuals are illustrated.

Figure 1: Example of an AS-OCT scan of an affected individual and three pedigrees seen in our family clinic (2006-2010) at Moorfields. Affected individuals are indicated as black squares or circles.
Sihota et al. found that shallower ACD, thicker lenses and shorter axial length (AL) segregated in family members affected with PACG compared to suspected and unaffected subjects [12]. Using the UBM, Etter et al. demonstrated a 50% rate of plateau iris configuration, in a small collection of predominantly white American families [15].
Previous heritability and segregation analyses
The most recent segregation analysis reporting 114 PACG families from Chongqing, China [16], found a significant difference between gender groups. They concluded that the inheritance of shallow angles may be a sex-influenced trait with a reported female:male ratio of 2.87:1, consistent with the ratio found in other East Asian population studies of PACG [4,17]. Families with an unaffected (U) parent and an affected parent (A) accorded with an autosomal dominant hereditary trait, with the highest heritability found for female relatives with female probands [16].
Several investigators have provided data on the segregation of ocular biometry and PACG. Tomlinson and Leighton [14] examined 16 index PACG patients, their relatives (seven siblings, 14 offspring) and 49 controls, finding that the unaffected relatives and patients with PACG also had smaller corneal diameters, shallower (ACDs), thicker crystalline lenses and shorter axial lengths compared to age-matched controls. However, those affected with PACG had more anteriorly positioned lenses, which were not observed in unaffected siblings and offspring. This finding was also observed by Rosengren [18] and Philips and Storey [19]. Törnquist from Sweden [20] found that eyes with PACG had ACDs of 1.0mm less than normal eyes, or two-thirds of the normal depth at corresponding ages.
Twin studies are often the starting point of dissecting genetic conditions, but there are no reported twin studies in PACG. A curious case report of two elderly identical twins who were in a heated argument and fist fight and both went into AAC together demonstrates that stress and other physiological triggers can influence disease presentation [21]. Hypermetropic refractive error is associated with angle-closure, but its contribution to PACG is likely to be incorporated with the heritability of ACD and AL [22-24].
Genetic linkage and candidate gene studies
Once a clear phenotype is identified, such as AAC or PACG, genetic studies can be performed. Aung et al. reported genetic linkage in two large Chinese families, examining the sharing of phenotypic and genetic factors, more statistically found together in affected individuals than unaffected ones [25]. To date, this method of genetic linkage has not identified any causative genes for PACG. However, in more extreme forms of the disease, two genes, membrane frizzled related protein (MFRP, associated with retinitis pigmentosa) [26,27] and protease, serine 56 (PRSS56) [28] have been identified to be causative for autosomal recessive microphthalmia / nanophthalmia. Both genes were identified using linkage analysis.
Aung et al. also examined 108 patients with PACG (49 with AAC and 59 chronic cases) for MFRP and CHX10 mutations [29]. CHX10 is a homeobox-containing transcription factor critical for progenitor cell proliferation and bipolar cell determination in the developing retina. One potential disease causing variant, G243D was observed in CHX10 in a patient with acute PACG who also had a MFRP R257H mutation. The clinical characteristics of the patient involved: an 82-year-old patient diagnosed at 75y with bilateral cup to disc ratio (CDR) of 0.8 and 360° of PAS and biometry of 21.05mm for AL and 2.09mm for ACD. This patient also carried an R46X polymorphism in the myocilin gene (MYOC) [30]. MYOC is an autosomal dominant genetic cause of POAG. The G243D mutation in CHX10 may be pathogenic as it was not found in 400 normal controls, but it was not possible to show segregation of the G243D mutation in the subject’s family members [29]. This was an example where sequence changes in three / multiple genes may each be contributing to the disease in an individual patient with late onset symptomatic PACG.
Genome wide association studies
PACG is clearly a complex disease with multifactorial inheritance, so it is no surprise that more success has been achieved with genome-wide association studies (GWAS). Using this methodology in 2012, three single-nucleotide polymorphisms (SNPs), rs11024102 in PLEKHA7, rs3753841 in COL11A1 (a connective tissue gene) and rs1015213 between PCMTD1 and ST18 were found to be associated with AAC/PACG [3]. Day et al. compared these three SNPs for association to ACD, AL and corneal keratometry using an additive genetic model for each allele in the EPIC-eyes cohort. The presence of one A allele for rs1015213 was associated with a 0.07mm shallowing of ACD compared to wild type homozygotes after adjusting for the effects of age and sex [31]. The other two SNPs were not associated with the biometric characteristics tested. Little is known about the function of PCMTD1 or ST18, the latter encodes for suppression of tumorigenecity 18 which is thought to modulate inflammation and apoptosis in fibroblasts [32] and has been reported as a breast cancer tumour suppressor gene [33].
The only GWAS study of ACD to date was reported by Vithana et al. where three well-characterised population based studies: Singapore Malay Eye Study (SiMES), Singapore Indian Eye Study (SINDI) and Beijing Eye Study (BES) with a total of 5308 participants were analysed. Two strongly associated intragenic SNPs on chromosome 3q27.1 were identified [34]. The gene of interest, ABCC5 (rs1401999) was widely expressed and regulates cGMP levels in its role as an organic anion pump. They were able to replicate the finding in a further cohort of Chinese subjects but not in Caucasians. This SNP was only marginally associated with PACG in a group of patients. Interestingly, this gene was not found to be replicated in the meta-analysis paper presented by Rong et al. in 2016 [1]. Nongpiur et al. hypothesise that the ABCC family of genes may be involved in glaucoma as ABCC4 has previously been found in the trabecular meshwork of a rabbit model with pigmentary glaucoma [34]. Perhaps overlapping anterior segment phenotypes will begin to unravel as more glaucoma genetic studies take place.
The most recent GWAS publication in PACG reported five further SNPS in EPDR1, CHAT, GLIS3, FERMT2, DPM2-FAM10A genes [35], and confirmed the original three loci (PLEKHA7, COL11A1 and PCTMD1-ST18) in the 2012 Vithana paper [3] to be associated with AAC and PACG.
Discussion
The strongest association signal to date is PLEKHA7. This cell-cell signaling gene lies close to the NNO1 locus linked to extreme nanophthalmos where axial lengths of less than 19mm and refractive errors of more than +7DS were observed [36]. The GWAS could not demonstrate an effect of PLEKHA7 on axial biometry or nanophthalmos. However, it does suggest that physiological mechanisms that maintain homeostasis in epithelial and endothelial tissues can alter the susceptibility to AAC and PACG, i.e. more advanced stages of the disease.
In GWAS studies, often the only SNPs that are reported are ones that have reached genome-wide statistical significance set at a threshold of P<5 x 10-8. In the 2012 Vithana paper [36], the fourth most significant locus was thioredoxin reductase 2 (TXNRD2), a gene involved in oxidative stress. Tissue expression studies and the relationship between this gene and its potential contribution to AAC or glaucoma have not yet been studied. The enzyme thioredoxin reductase (TR) is a dimeric NADPH-dependent FAD containing enzyme that catalyses the reduction of the active site of disulphide of thioredoxin and other substrates. It is a member of a family of pyridine nucleotide-disulphide oxidoreductases and a key enzyme in the regulation of the intracellular redox environment both in the cytosol and mitochondria [3]. This gene partially overlaps with the COMT gene on chromosome 22. The COMT gene encodes for catechol-O-methyltransferase (COMT) which catalyses the transfer of a methyl group from S-adenosylmethionine to catecholamines, including the neurotransmitters dopamine, epinephrine and norepinephrine. Oestrogens such as 17β-oestradiol, catabolised in hydroxylation reactions, are inactivated by methylation, a process catalysed by COMT [38]. This pathway is important in oestradiol metabolism.
There is increasing evidence that the inheritance of particular variants in these oestrogen-metabolising genes can modulate the risk of hormone dependent disorders such as prostate, ovarian, lung and breast cancer [38]. Given the female preponderance to ACG, this area of work to understand the molecular genetic and biochemical mechanisms underlying PACG could lead to breakthroughs in our scientific understanding and delivery of novel therapies.
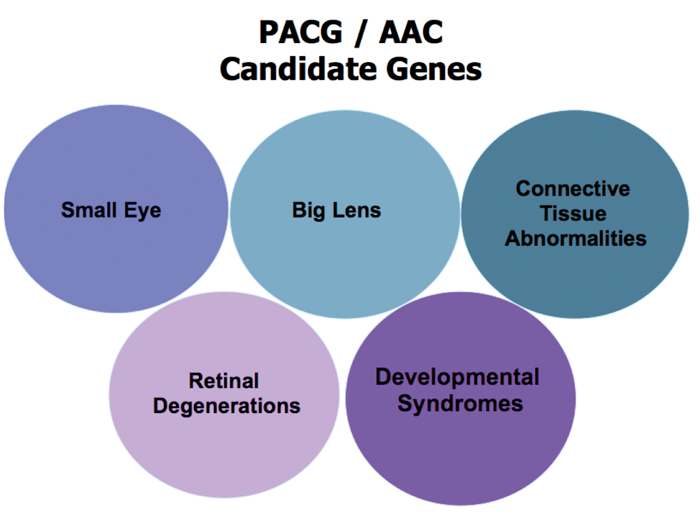
Figure 2: Phenotypes to look out for when considering a genetic cause for angle closure glaucoma.
Conclusion
There is emerging research in the genetics of ACG and keen phenotyping (skilled clinicians) in partnership with molecular biologists and statisticians can bridge the gaps in our knowledge of the disease. Figure 2 simplifies the phenotypes that have been discussed in this article. Even without genetic data, ophthalmic practitioners can recommend screening of family members, and proactively teach and learn gonioscopy, as accurate elucidation of clinical signs and appropriate management is essential for the diagnosis and prognosis of patients with AAC and PACG.
References
1. Rong SS, Tang FY, Chu WK, et al. Genetic associations of primary angle-closure disease: a systematic review and meta-analysis. Ophthalmology 2016;123(6):1211-21.
2. Foster PJ, Buhrmann RR, Quigley HA, Johnson GJ. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol 2002;86:238-42.
3. Vithana EN, Khor CC, Qiao C, et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat Genet 2012;44:1142-6.
4. Foster PJ, Oen FT, Machin D, et al. The prevalence of glaucoma in Chinese residents of Singapore: a cross-sectional population survey of the Tanjong Pagar district. Arch Ophthalmol 2000;118:1105-11.
5. Merula RV, Cronemberger S, Calixto N. [Incidence of primary angle-closure glaucoma in the Glaucoma Service of the Sao Geraldo Hospital]. Arq Bras Oftalmol 2008;71:389-93.
6. Qu W, Li Y, Song W, et al. Prevalence and risk factors for angle-closure disease in a rural Northeast China population: a population-based survey in Bin County, Harbin. Acta Ophthalmol 2001;89:e515-20.
7. Amerasinghe N, Zhang J, Thalamuthu A, et al. The heritability and sibling risk of angle closure in Asians. Ophthalmology 2011;118:480-5.
8. Kong X, Chen Y, Chen X, Sun X. Influence of family history as a risk factor on primary angle closure and primary open angle glaucoma in a Chinese population. Ophthalmic Epidemiol 2011;18:226-32.
9. Kavitha S, Zebardast N, Palaniswamy K, et al. Family history is a strong risk factor for prevalent angle closure in a South Indian population. Ophthalmology 2014;121:2091-7.
10. Lowe RF. Primary angle-closure glaucoma. Family histories and anterior chamber depths. Br J Ophthalmol 1964;48:191-5.
11. Lowe RF. Primary angle-closure glaucoma. Inheritance and environment. Br J Ophthalmol 1972;56:13-20.
12. Sihota R, Ghate D, Mohan S, et al. Study of biometric parameters in family members of primary angle closure glaucoma patients. Eye 2008;22:521-7.
13. Spaeth GL. Gonioscopy: uses old and new. The inheritance of occludable angles. Ophthalmology 1978;85:222-32.
14. Tomlinson A, Leighton DA. Ocular dimensions in the heredity of angle-closure glaucoma. Br J Ophthalmol 1973;57:475-86.
15. Etter JR, Affel EL, Rhee DJ. High prevalence of plateau iris configuration in family members of patients with plateau iris syndrome. J Glaucoma 2006;15:394-8.
16. Tu YS, Yin ZQ, Pen HM, Yuan CM. Genetic heritability of a shallow anterior chamber in Chinese families with primary angle closure glaucoma. Ophthalmic Genet 2008;29:171-6.
17. Foster PJ, Baasanhu J, Alsbirk PH, et al. Glaucoma in Mongolia. A population-based survey in Hovsgol province, northern Mongolia. Arch Ophthalmol 1996;114:1235-41.
18. Rosengren B. Studies in depth of the anterior chamber of the eye in primary glaucoma. AMA Arch Ophthalmol 1950;44:523-38.
19. Phillips CI, Storey JK. Glaucoma geometry. Exp Eye Res 1971;11:140-1.
20. Törnquist R. Shallow anterior chamber in acute glaucoma; a clinical and genetic study. Acta Ophthalmol Suppl 1953;39:1-74.
21. Talluto D, Feith M, Allee S. Simultaneous angle closure in twins. J Glaucoma 1998;7:68-9.
22. Congdon N, Wang F, Tielsch JM. Issues in the epidemiology and population-based screening of primary angle-closure glaucoma. Surv Ophthalmol 1992;36(6):411-23.
23. Wickremasinghe S. Foster PJ, Uranchimeg D, et al. Ocular biometry and refraction in Mongolian adults. Invest Ophthalmol Vis Sci 2004;45:776-83.
24. Wong TY, Foster PJ, Johnson GJ, et al. The relationship between ocular dimensions and refraction with adult stature: the Tanjong Pagar Survey. Invest Ophthalmol Vis Sci 2001;42:1237-42.
25. Aung T, Bowman R, Chew PT, et al. Genome-wide linkage scan for primary angle-closure glaucoma. Invest Ophthalmol Vis Sci 2003;44:3224.
26. Sundin OH, Dharmaraj S, Bhutto IA, et al. Developmental basis of nanophthalmos: MFRP is required for both prenatal ocular growth and postnatal emmetropization. Ophthalmic Genet 2008;29:1-9.
27. Ayala-Ramirez R, Graue-Wiechers F, Robredo V, et al. A new autosomal recessive syndrome consisting of posterior microphthalmos, retinitis pigmentosa, foveoschisis, and optic disc drusen is caused by a MFRP gene mutation. Mol Vis 2006;12:1483-9.
28. Nair KS, Hmani-Aifa M, Ali Z, et al. Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat Genet 2011;43:579-84.
29. Aung T. Lim MC, Wong TT, et al. Molecular analysis of CHX10 and MFRP in Chinese subjects with primary angle closure glaucoma and short axial length eyes. Mol Vis, 2008;14:1313-8.
30. Aung T, Yong VH, Chew PT, et al. Molecular analysis of the myocilin gene in Chinese subjects with chronic primary-angle closure glaucoma. Invest Ophthalmol Vis Sci 2005;46:1303-6.
31. Day AC, Luben R, Khawaja AP, et al. Genotype-phenotype analysis of SNPs associated with primary angle closure glaucoma (rs1015213, rs3753841 and rs11024102) and ocular biometry in the EPIC-Norfolk Eye Study. Br J Ophthalmol 2013;97(6):704-7.
32. Yang J, Siqueira MF, Behl Y, et al. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J 2008;22:3956-67.
33. Jandrig B, Seitz S, Hinzmann B, et al. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene 2004;23:9295-302.
34. Nongpiur ME, Khor CC, Jia H, et al. ABCC5, a gene that influences the anterior chamber depth, is associated with primary angle closure glaucoma. PLoS Genet 2014;10:e1004089.
35. Khor CC, Do T, Jia H, et al. Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nat Genet 2016;48:556-62.
36. Othman MI, Sullivan SA, Skuta GL, et al. Autosomal dominant nanophthalmos (NNO1) with high hyperopia and angle-closure glaucoma maps to chromosome 11. Am J Hum Genet 1998;63:1411-8.
37. Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 2000;267:6102-9.
38. Huber JC, Schneeberger C, Tempfer CB. Genetic modeling of estrogen metabolism as a risk factor of hormone-dependent disorders. Maturitas 2002;41(Suppl 1):S55-64.
Further reading
Contact your local Allergan and ATHENA (Allergan Ophthalmic Professionals Educational Alliance) representative for the Early Diagnosis Program 4 – Gonioscopy in the diagnosis of glaucoma by Sancy Low and Gus Gazzard to get teaching materials and arrange departmental gonioscopy learning.
Take home message
-
Genetic aetiologies are different for PACG and POAG.
-
Older female first degree relatives are at highest risk of developing disease, with small ocular biometry and physical size as key contributing factors.
-
Glaucoma and gonioscopic examinations can be recommended to patients who have AAC or PACG.
-
Hyperopic refractive error overlaps with PACG, and may have shared genetic mechanisms.
-
When a patient with atypical ACG presents to your clinic, consider connective tissue disorders.
Declaration of competing interests: None declared.
COMMENTS ARE WELCOME